Learning to DANNCE
By Jonathan McCall, MS, and Rabail Baig, MA
A group of neuroscientists and machine learning experts are developing new ways to analyze animal movement and behavior to gain insights into the inner workings of the nervous system.
It’s no secret that you can guess what an animal is thinking or feeling based on the way it moves. The dog that wags its tail is happy to see you; bared teeth and bristling fur in the same animal convey something quite different.
But what if we could go far beyond these relatively obvious interpretations of animal behavior? What if we could learn to decode these eloquent movements and translate them into a detailed understanding of the inner workings of an animal’s brain and body? Could we then learn to apply these techniques to the human brain, unlocking new insights into states of health and disease?
These questions are at the heart of a study undertaken by a team of researchers from Duke University, Harvard, MIT, Rockefeller University, and Columbia University. Combining expertise from the disciplines of neurobiology and artificial intelligence, they developed a system that captures detailed, multiple-view video of animals in their natural environment, and then uses data from those video images to build a detailed model of how the animal moves. This allows scientists to use movement and behavior as a window into brain function.
“There has been this huge technological gap that’s been hampering our ability to understand the brain in health and disease,” says Timothy Dunn, PhD, an assistant professor with the Duke University Department of Neurosurgery, a researcher with Duke AI Health, and a member of the study team. Dunn, who specializes in the application of machine learning in neuroscience, notes that while there has been significant progress in directly measuring and manipulating brain activity, methods that allow scientists to quantify and measure the output of that activity– in this case, movement and behavior—have lagged behind. It was this gap that Dunn and his team were attempting to fill.

One major driver for the group’s efforts was the desire to use analysis of animal behavior to shed light on Parkinson disease, autism, and other disorders where highly detailed characterization of movement might reveal additional information about the disease and how it manifests—and possibly suggest new avenues for therapeutic intervention.
Seeing Deeper with Machine Learning
“In animals where we’d like to be able to study the mechanisms underlying these disorders, or the potential effects of therapies, being unable to measure the actual symptoms is hugely limiting,” Dunn notes. “We need to be able to measure all of these variables that could be controlled by the brain and could be affected by mutations, could be affected by drugs and therapies—and that hasn’t been possible until we set out to build our new technique.”
“Ultimately, science is measurement,” says Dunn, “and if we can’t measure and quantify key components of a system rigorously and without bias, then it’s difficult to describe the work as science.”
Analyzing video of animal behavior to glean insights into neurological states is not itself a new approach. However, currently available methods have substantial limitations.
“In freely moving animals, state-of-the-art technology only allows us to measure how much the animal is moving, or how fast the animal is turning to the left or turning to the right, or how fast it is moving over some space in a behavioral arena,” says Dunn, who is the lead author on a paper published this week in Nature Methods that describes the results from the group’s most recent work.
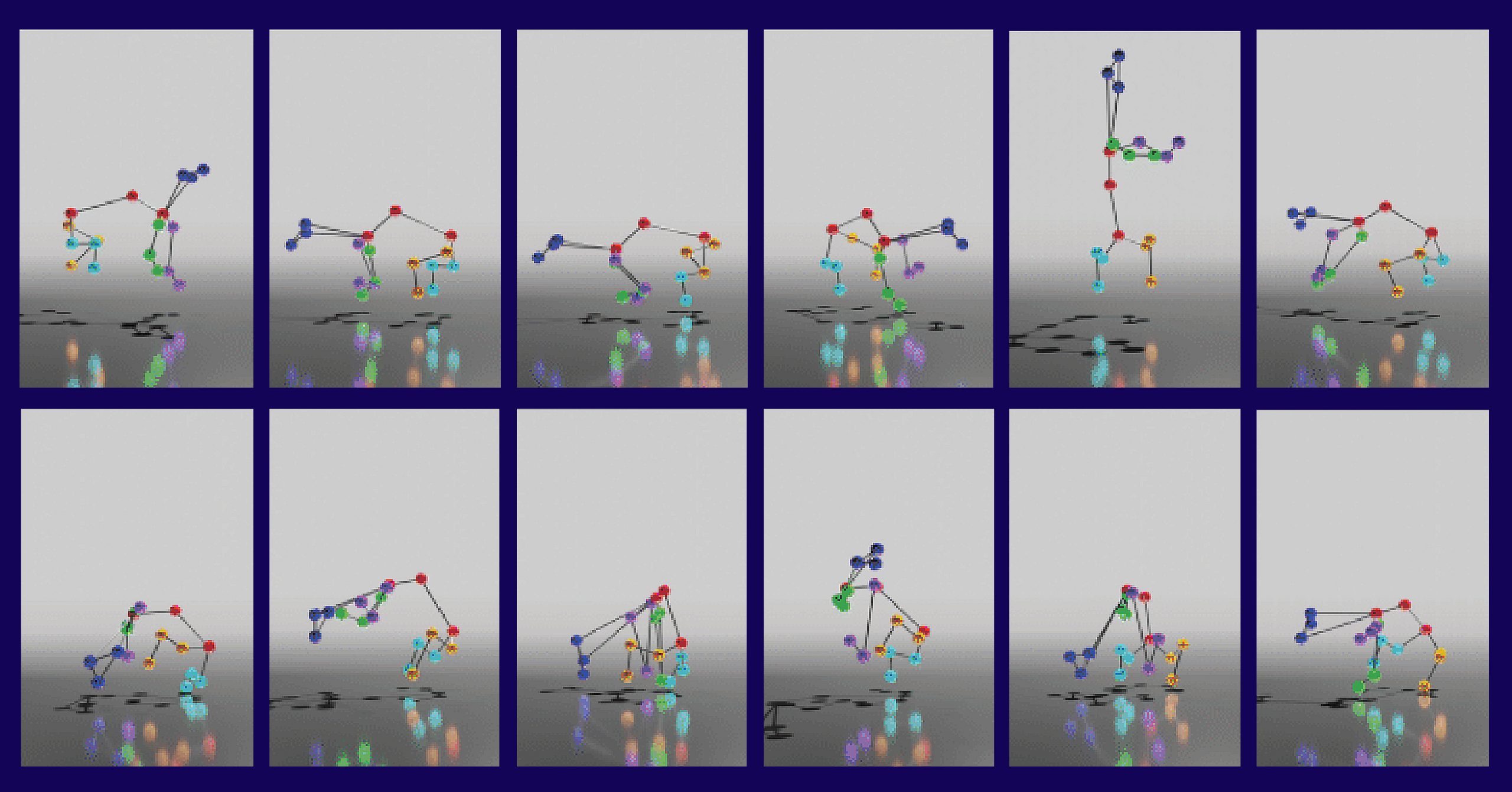
This need to capture fine-grained details spurred the creation of DANNCE (short for 3-Dimensional Aligned Neural Network for Computational Ethology), which uses a kind of artificial intelligence called a convolutional neural network to learn from millions of video frames how animals—in this case, rats—move in a natural setting. As the system was trained on images of different well-defined poses, it was increasingly able to identify the positions of different body parts, and from there, to use information from multiple camera angles to make inferences about how the animal was moving in three-dimensional space.
Dunn describes the system as being superficially similar to motion capture, a computer animation technology widely used for tracking the movements of human actors and then superimposing computer-animated features on top. But unlike motion capture, DANNCE does not require large, expensive camera systems, nor does it require the animal to be equipped with wearable markers. Most importantly, it can be used in an animal’s normal environment—and it works even when the observer’s view of the animal is partially obscured, according to senior author Bence P. Ölveczky, PhD, a professor and researcher with Harvard University’s Department of Organismic and Evolutionary Biology.
“What’s remarkable is that this little network now has its own secrets and can infer the precise movements of animals it wasn’t trained on, even when large parts of their body is hidden from view,” notes Ölveczky.
This capacity to fill in gaps even when limited to a view from a single camera is a crucial part of what sets DANNCE apart from existing methods.
“We compared DANNCE to other networks designed to do similar tasks and found DANNCE outperformed them,” said Jesse Marshall, PhD, a Harvard postdoctoral researcher who co-led the study with Dunn.
Next Steps for DANNCE
DANNCE was developed using rats as a model animal, but it can be applied to other species.
“Rats, mice, marmosets, black-capped chickadees,” adds Dunn, listing the various animals in which the system has already been tested. But while DANNCE can be adapted for use in other animals and even in humans, this requires fine-tuning the system with additional examples—and the more examples the neural net has to learn from, the more finely its predictive capabilities are honed. Dunn points out that the current study builds on findings from an earlier investigation that applied painstaking analysis of animal movement to reveal subtle but meaningful difference. He explains that when the 3D movement model was applied to rats being given low doses of caffeine or amphetamines, the more detailed measurements revealed substantial differences in the animals’ responses to the two stimulants, which previously had been opaque to earlier methods of detection.
The techniques the group is developing could have direct applications to the clinic, one of which is that increasingly precise classification of movements may reveal subtle variations in disease or differences in underlying mechanisms, leading to highly targeted interventions. Dunn notes that the team’s efforts can be described as an attempt to develop a quantitative language that opens a new window into the function of complex processes implemented in the brain, and allows those functions to precisely measured and described.
“Ultimately, science is measurement,” says Dunn, “and if we can’t measure and quantify key components of a system rigorously and without bias, then it’s difficult to describe the work as science.”
In addition to Dunn, Marshall, and Ölveczky, study authors include Kyle S. Severson, Diego E. Aldarondo, David G. C. Hildebrand, Selmaan N. Chettih, William L. Wang, Amanda J. Gellis, David E. Carlson, Dmitriy Aronov, Winrich A. Freiwald, and Fan Wang.
Geometric deep learning enables 3D kinematic profiling across species and environments. Nature Methods. DOI: 10.1038/s41592-021-01106-6
Acknowledgments
With thanks to Wendy Heywood from the Harvard University Department of Organismic and Evolutionary Biology for quotes from Drs. Ölveczky and Marshall. We also thank Duke Research’s Veronique Koch and Karl Bates for video editing and additional support.
In this video interview, Timothy Dunn provides some additional insights into the DANNCE study
